Protein-coding gene in the species Homo sapiens
| LILRB1 |
|---|
 |
| Available structures |
|---|
| PDB | Human UniProt search: PDBe RCSB |
|---|
| List of PDB id codes |
|---|
1G0X, 1P7Q, 1UFU, 1UGN, 1VDG, 3D2U, 4LL9, 4NO0 |
|
|
| Identifiers |
|---|
| Aliases | LILRB1, CD85J, ILT-2, ILT2, LIR-1, LIR1, MIR-7, MIR7, PIR-B, PIRB, leukocyte immunoglobulin like receptor B1 |
|---|
| External IDs | OMIM: 604811 HomoloGene: 88463 GeneCards: LILRB1 |
|---|
| Gene location (Human) |
|---|
 | | Chr. | Chromosome 19 (human)[1] |
|---|
| | Band | 19q13.42 | Start | 54,617,158 bp[1] |
|---|
| End | 54,638,022 bp[1] |
|---|
|
| RNA expression pattern |
|---|
| Bgee | | Human | Mouse (ortholog) |
|---|
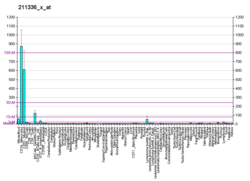
| Top expressed in | - spleen
- monocyte
- appendix
- blood
- lymph node
- bone marrow cells
- upper lobe of left lung
- right lung
- superficial temporal artery
- right coronary artery
|
| | | More reference expression data |
|
|---|
| BioGPS | 
 | | More reference expression data |
|
|---|
|
| Gene ontology |
|---|
| Molecular function | - protein homodimerization activity
- MHC class I receptor activity
- SH2 domain binding
- protein phosphatase 1 binding
- HLA-A specific inhibitory MHC class I receptor activity
- HLA-B specific inhibitory MHC class I receptor activity
- MHC class I protein binding
- amyloid-beta binding
- protein binding
- MHC class Ib receptor activity
- MHC class Ib protein complex binding
- MHC class Ib protein binding
| | Cellular component | - cytoplasm
- integral component of membrane
- membrane
- plasma membrane
- extracellular region
- external side of plasma membrane
| | Biological process | - negative regulation of endocytosis
- dendritic cell differentiation
- negative regulation of mononuclear cell proliferation
- negative regulation of interferon-gamma production
- adaptive immune response
- Fc receptor mediated inhibitory signaling pathway
- negative regulation of osteoclast development
- immune system process
- response to virus
- receptor internalization
- negative regulation of cell cycle
- negative regulation of cytokine production involved in immune response
- negative regulation of serotonin secretion
- T cell proliferation involved in immune response
- negative regulation of alpha-beta T cell activation
- negative regulation of T cell mediated cytotoxicity
- positive regulation of cytolysis
- negative regulation of T cell proliferation
- positive regulation of gene expression
- defense response to virus
- negative regulation of calcium ion transport
- regulation of immune response
- positive regulation of apoptotic process
- negative regulation of natural killer cell mediated cytotoxicity
- negative regulation of T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell
- cellular response to lipopolysaccharide
- immune response-inhibiting cell surface receptor signaling pathway
- positive regulation of gamma-delta T cell activation involved in immune response
- negative regulation of dendritic cell apoptotic process
- negative regulation of dendritic cell differentiation
- signal transduction
- positive regulation of transcription by RNA polymerase II
- negative regulation of CD8-positive, alpha-beta T cell activation
- positive regulation of defense response to virus by host
- positive regulation of macrophage cytokine production
| | Sources:Amigo / QuickGO |
|
| Orthologs |
|---|
| Species | Human | Mouse |
|---|
| Entrez | | |
|---|
| Ensembl | ENSG00000104972
ENSG00000276452
ENSG00000277134
ENSG00000274669
ENSG00000277807
|
|---|
n/a |
| |
|---|
| UniProt | | |
|---|
| RefSeq (mRNA) | NM_001081637
NM_001081638
NM_001081639
NM_001278398
NM_001278399
|
|---|
NM_006669
NM_001388355
NM_001388356
NM_001388357
NM_001388358 |
| |
|---|
| RefSeq (protein) | NP_001075106
NP_001075107
NP_001075108
NP_001265327
NP_001265328
|
|---|
NP_006660 |
| |
|---|
| Location (UCSC) | Chr 19: 54.62 – 54.64 Mb | n/a |
|---|
| PubMed search | [2] | n/a |
|---|
|
| Wikidata |
|
Leukocyte immunoglobulin-like receptor subfamily B member 1 is a protein that in humans is encoded by the LILRB1 gene.[3][4]
Function
This gene is a member of the leukocyte immunoglobulin-like receptor (LIR) family, which is found in a gene cluster at chromosomal region 19q13.4. The encoded protein belongs to the subfamily B class of LIR receptors which contain two or four extracellular immunoglobulin domains, a transmembrane domain, and two to four cytoplasmic immunoreceptor tyrosine-based inhibitory motifs (ITIMs). The receptor is expressed on immune cells where it binds to MHC class I molecules on antigen-presenting cells and transduces a negative signal that inhibits stimulation of an immune response. It is thought to control inflammatory responses and cytotoxicity to help focus the immune response and limit autoreactivity. Multiple transcript variants encoding different isoforms have been found for this gene.[5]
See also
References
- ^ a b c ENSG00000276452, ENSG00000277134, ENSG00000274669, ENSG00000277807 GRCh38: Ensembl release 89: ENSG00000104972, ENSG00000276452, ENSG00000277134, ENSG00000274669, ENSG00000277807 – Ensembl, May 2017
- ^ "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ^ Cosman D, Fanger N, Borges L, Kubin M, Chin W, Peterson L, Hsu ML (Aug 1997). "A novel immunoglobulin superfamily receptor for cellular and viral MHC class I molecules". Immunity. 7 (2): 273–82. doi:10.1016/S1074-7613(00)80529-4. PMID 9285411.
- ^ Colonna M, Navarro F, Bellón T, Llano M, García P, Samaridis J, Angman L, Cella M, López-Botet M (Dec 1997). "A common inhibitory receptor for major histocompatibility complex class I molecules on human lymphoid and myelomonocytic cells". The Journal of Experimental Medicine. 186 (11): 1809–18. doi:10.1084/jem.186.11.1809. PMC 2199153. PMID 9382880.
- ^ "Entrez Gene: LILRB1 leukocyte immunoglobulin-like receptor, subfamily B (with TM and ITIM domains), member 1".
Further reading
- Maruyama K, Sugano S (Jan 1994). "Oligo-capping: a simple method to replace the cap structure of eukaryotic mRNAs with oligoribonucleotides". Gene. 138 (1–2): 171–4. doi:10.1016/0378-1119(94)90802-8. PMID 8125298.
- Hirohashi N, Nakao M, Kubo K, Yamada A, Shichijo S, Hara A, Sagawa K, Itoh K (Dec 1993). "A novel antigen (H47 Ag) on human lymphocytes involved in T cell activation". Cellular Immunology. 152 (2): 371–82. doi:10.1006/cimm.1993.1298. PMID 8258145.
- Samaridis J, Colonna M (Mar 1997). "Cloning of novel immunoglobulin superfamily receptors expressed on human myeloid and lymphoid cells: structural evidence for new stimulatory and inhibitory pathways". European Journal of Immunology. 27 (3): 660–5. doi:10.1002/eji.1830270313. PMID 9079806. S2CID 2212182.
- Wagtmann N, Rojo S, Eichler E, Mohrenweiser H, Long EO (Aug 1997). "A new human gene complex encoding the killer cell inhibitory receptors and related monocyte/macrophage receptors". Current Biology. 7 (8): 615–8. Bibcode:1997CBio....7..615W. doi:10.1016/S0960-9822(06)00263-6. PMID 9259559. S2CID 14484039.
- Suzuki Y, Yoshitomo-Nakagawa K, Maruyama K, Suyama A, Sugano S (Oct 1997). "Construction and characterization of a full length-enriched and a 5'-end-enriched cDNA library". Gene. 200 (1–2): 149–56. doi:10.1016/S0378-1119(97)00411-3. PMID 9373149.
- Fanger NA, Cosman D, Peterson L, Braddy SC, Maliszewski CR, Borges L (Nov 1998). "The MHC class I binding proteins LIR-1 and LIR-2 inhibit Fc receptor-mediated signaling in monocytes". European Journal of Immunology. 28 (11): 3423–34. doi:10.1002/(SICI)1521-4141(199811)28:11<3423::AID-IMMU3423>3.0.CO;2-2. PMID 9842885.
- Navarro F, Llano M, Bellón T, Colonna M, Geraghty DE, López-Botet M (Jan 1999). "The ILT2(LIR1) and CD94/NKG2A NK cell receptors respectively recognize HLA-G1 and HLA-E molecules co-expressed on target cells". European Journal of Immunology. 29 (1): 277–83. doi:10.1002/(SICI)1521-4141(199901)29:01<277::AID-IMMU277>3.0.CO;2-4. PMID 9933109.
- Chapman TL, Heikeman AP, Bjorkman PJ (Nov 1999). "The inhibitory receptor LIR-1 uses a common binding interaction to recognize class I MHC molecules and the viral homolog UL18". Immunity. 11 (5): 603–13. doi:10.1016/S1074-7613(00)80135-1. PMID 10591185.
- Liu WR, Kim J, Nwankwo C, Ashworth LK, Arm JP (Jul 2000). "Genomic organization of the human leukocyte immunoglobulin-like receptors within the leukocyte receptor complex on chromosome 19q13.4". Immunogenetics. 51 (8–9): 659–69. doi:10.1007/s002510000183. PMID 10941837. S2CID 23687483.
- Chapman TL, Heikema AP, West AP, Bjorkman PJ (Nov 2000). "Crystal structure and ligand binding properties of the D1D2 region of the inhibitory receptor LIR-1 (ILT2)". Immunity. 13 (5): 727–36. doi:10.1016/S1074-7613(00)00071-6. PMID 11114384.
- Lepin EJ, Bastin JM, Allan DS, Roncador G, Braud VM, Mason DY, van der Merwe PA, McMichael AJ, Bell JI, Powis SH, O'Callaghan CA (Dec 2000). "Functional characterization of HLA-F and binding of HLA-F tetramers to ILT2 and ILT4 receptors". European Journal of Immunology. 30 (12): 3552–61. doi:10.1002/1521-4141(200012)30:12<3552::AID-IMMU3552>3.0.CO;2-L. PMID 11169396. S2CID 42462661.
- Young NT, Canavez F, Uhrberg M, Shum BP, Parham P (2001). "Conserved organization of the ILT/LIR gene family within the polymorphic human leukocyte receptor complex". Immunogenetics. 53 (4): 270–8. doi:10.1007/s002510100332. PMID 11491530. S2CID 2819935.
- Saverino D, Merlo A, Bruno S, Pistoia V, Grossi CE, Ciccone E (Jan 2002). "Dual effect of CD85/leukocyte Ig-like receptor-1/Ig-like transcript 2 and CD152 (CTLA-4) on cytokine production by antigen-stimulated human T cells". Journal of Immunology. 168 (1): 207–15. doi:10.4049/jimmunol.168.1.207. PMID 11751964.
- Bellón T, Kitzig F, Sayós J, López-Botet M (Apr 2002). "Mutational analysis of immunoreceptor tyrosine-based inhibition motifs of the Ig-like transcript 2 (CD85j) leukocyte receptor". Journal of Immunology. 168 (7): 3351–9. doi:10.4049/jimmunol.168.7.3351. PMID 11907092.
- Nikolova M, Musette P, Bagot M, Boumsell L, Bensussan A (Aug 2002). "Engagement of ILT2/CD85j in Sézary syndrome cells inhibits their CD3/TCR signaling". Blood. 100 (3): 1019–25. doi:10.1182/blood-2001-12-0303. PMID 12130517. S2CID 7351536.
- Willcox BE, Thomas LM, Chapman TL, Heikema AP, West AP, Bjorkman PJ (Oct 2002). "Crystal structure of LIR-2 (ILT4) at 1.8 A: differences from LIR-1 (ILT2) in regions implicated in the binding of the Human Cytomegalovirus class I MHC homolog UL18". BMC Structural Biology. 2: 6. doi:10.1186/1472-6807-2-6. PMC 130215. PMID 12390682.
- Achdout H, Arnon TI, Markel G, Gonen-Gross T, Katz G, Lieberman N, Gazit R, Joseph A, Kedar E, Mandelboim O (Jul 2003). "Enhanced recognition of human NK receptors after influenza virus infection". Journal of Immunology. 171 (2): 915–23. doi:10.4049/jimmunol.171.2.915. PMID 12847262.
External links
 | This membrane protein–related article is a stub. You can help Wikipedia by expanding it. |
This article incorporates text from the United States National Library of Medicine, which is in the public domain.
PDB gallery |
|---|
-
1g0x: CRYSTAL STRUCTURE OF THE LIGAND BINDING DOMAIN OF LIR-1 (ILT2) -
1p7q: Crystal Structure of HLA-A2 Bound to LIR-1, a Host and Viral MHC Receptor -
1ufu: Crystal structure of ligand binding domain of immunoglobulin-like transcript 2 (ILT2; LIR-1) -
1ugn: Crystal structure of LIR1.02, one of the alleles of LIR1 -
1vdg: Crystal structure of LIR1.01, one of the alleles of LIR1 |
|
|---|
| 1–50 | |
|---|
| 51–100 | |
|---|
| 101–150 | |
|---|
| 151–200 | |
|---|
| 201–250 | |
|---|
| 251–300 | |
|---|
| 301–350 | |
|---|